You can assign an individual deff for each propagation step and each element in x and y. Note there is a different nonlinear coefficient for eoo, oeo, ooe type (deff1) and eeo,eoe,oee type (deff2) nonlinear interaction types.
To implement a constant or a chirped poling period simply use a vector with Nz elements for the initialization of the deff. Each element of the vector defines the nonlinear coefficient used for a certain propagation step in the crystal, respectively.
L=obj.simProp.propagationLength;
z=linspace(obj.constVars.dz,L,obj.simProp.Nz);
PPmin=21.5e-6;
PPmax=21.5e-6;
deff=16e-12.*(round((sin(2*pi * ( 1/PPmin*z + (1/PPmax-1/PPmin)/L/2*z.^2)) +1 )/2)*2-1);
obj( 'deff1',deff);
To simulate a fan-out structured PPLN you can define a 3 dimensional deff-matrix with the dimensions deff[Nx,Ny,Nz], e.g.: (not tested)
polingPeriod=21.5e-6;
fanOut= 8e-6; %maxPoling-minPoling
polingWidth=12e-3; %spatial dimension in x direction
z=meshgrid(linspace(0,obj.simProp.propagationLength,obj.simProp.Nz),1:obj.simProp.Nx);
pol=meshgrid(fanOut/polingWidth * obj.constVect.x + polingPeriod , 1:obj.simProp.Nz)';
deff = 16e-12.*(round((sin(2.*pi./pol.*z)+1)/2)*2-1); % fan out poling structur in 1+1 dimensions -> array with (Nx,Nz) elements
if(Ny>1)
deff = permute(reshape(ndgrid(deff,1,1:obj.simProp.Ny),obj.simProp.Nx,obj.simProp.Nz,obj.simProp.Ny),[1,3,2]); % fan out poling structur in x-direction in 3+1 dimensions with (Nx,Ny,Nz) elements
end
obj( 'deff1',deff);
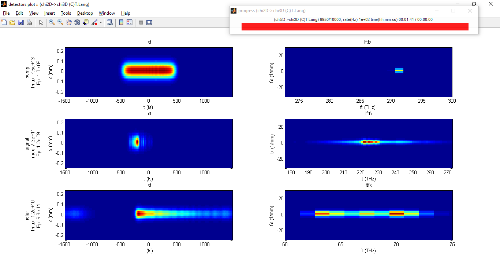
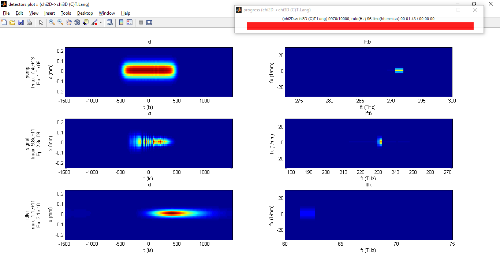
To give a rather academic example for a DFG in a chirped PPLN pumped at 1030 nm and broadband seeded at 1300 nm:
%create a new instance of chi3D
obj = chi3D('tag','PPLN example');
%set computational parameters
obj( 'Nx',32,'xWindow',0.0005,...
'Ny',1 ,'yWindow',0.0005,... %use Ny=1 for 2+1D, Ny=2^N for 3+1d
'Nt',2048, 'tWindow',3e-12,'shiftFreqWindow',0,...
'Nz',10000, 'propagationLength', 0.005,...
'GPUOn',0, 'abortIntensity', inf,...
'showResultEach',1000, 'showProgress',1,...
'material','LNcMgO','theta',1.5708,'phi',0,'plane','XZ');
% compute the deff-vector
L=obj.simProp.propagationLength;
z=linspace(obj.constVars.dz,L,obj.simProp.Nz);
PPmin=21.5e-6;
PPmax=23.5e-6;
deff=16e-12.*(round((sin(2*pi * ( 1/PPmin*z + (1/PPmax-1/PPmin)/L/2*z.^2)) +1 )/2)*2-1); % simple chirp between PPmin and PPmax
obj( 'deff1',deff,'eoo',0,'oeo',0,'ooo',1,...
'deff2',0 ,'oee',0,'eeo',0,'eee',0);
%input pulse definition
obj({ 'pulse1','EorI',1e-06,'polarization','o','centerWavelength',1.03e-06,...
'pulseDuration',6e-13,'beamShape_t',{'supergauss',10},...
'beamRadius_x',0.0001,'beamRadius_y',0.0001,... %please choose a reasonable values in y
});
obj({ 'pulse2','EorI',1e-09,'polarization','o','centerWavelength',1.307e-06,...
'pulseDuration',1e-14,'GD',2.5e-13,...
'beamRadius_x',0.0001,'beamRadius_y',0.0001,... %please choose a reasonable values in y
});
%define detectors
obj.detectors = rmfield(obj.detectors,fieldnames(obj.detectors)); %delete default detectors
obj.detectors.pump.plotIntegratedProfiles=1;
obj.detectors.pump.polarization='o';
obj.detectors.pump.ftlim=[3e8/1.1e-6,3e8/1e-6];
obj.detectors.signal.plotIntegratedProfiles=1;
obj.detectors.signal.polarization='o';
obj.detectors.signal.ftlim=[3e8/1.7e-6,3e8/1.1e-6];
obj.detectors.idler.plotIntegratedProfiles=1;
obj.detectors.idler.polarization='o';
obj.detectors.idler.ftlim=[3e8/5e-6,3e8/4e-6];
%run simulation
[E_ftfxfy_o,E_ftfxfy_e]=run(obj,{'pulse1'},{'pulse2'});
chirped
unchirped
Tino Lang